
Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics
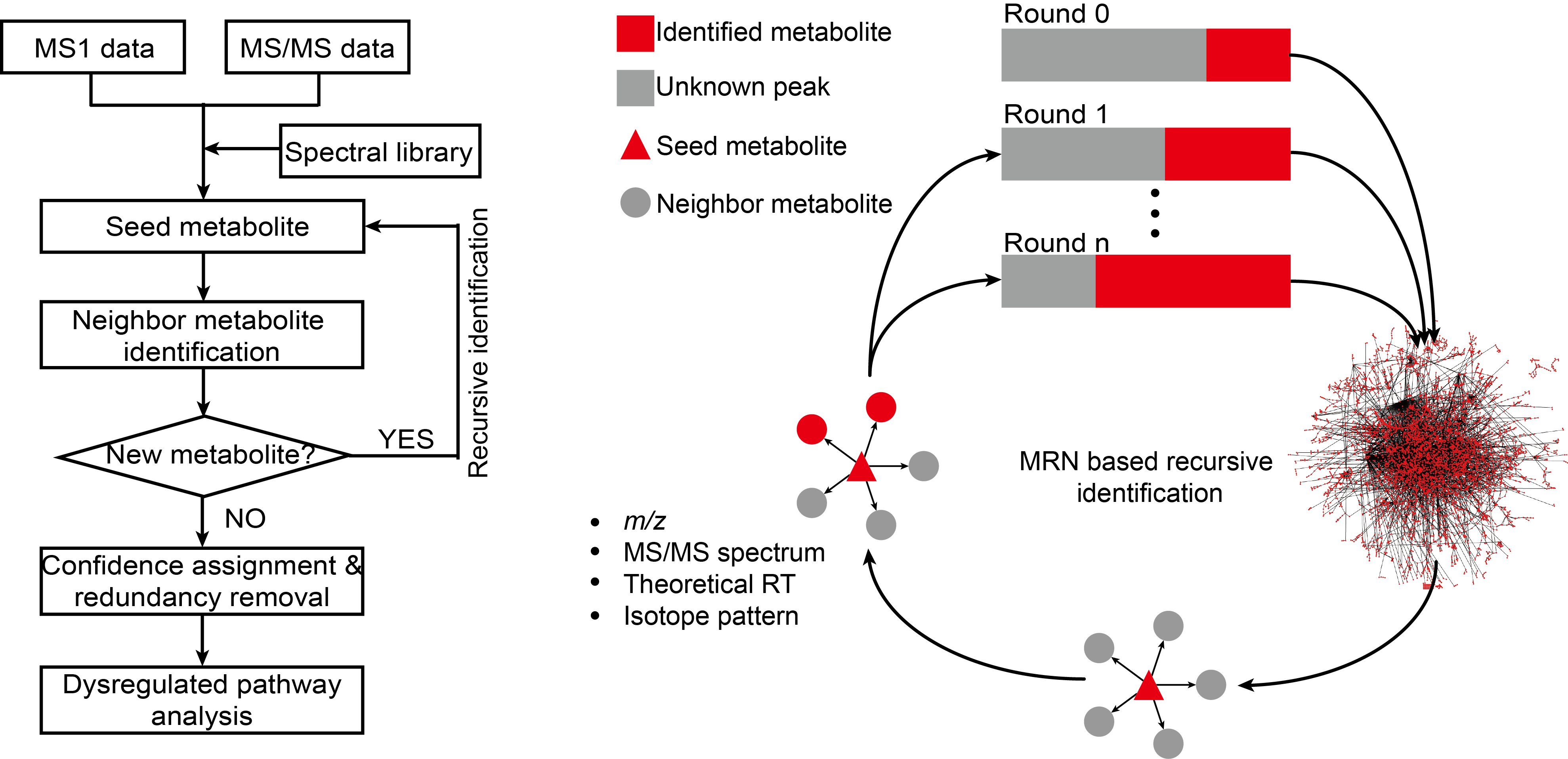
Introduction: Large-scale metabolite annotation is a challenge in liquid chromatogram-mass spectrometry (LC-MS)-based untargeted metabolomics. Here, we develop a metabolic reaction network (MRN)-based recursive algorithm (MetDNA) that expands metabolite annotations without the need for a comprehensive standard spectral library. MetDNA is based on the rationale that seed metabolites and their reaction-paired neighbors tend to share structural similarities resulting in similar MS2 spectra. MetDNA characterizes initial seed metabolites using a small library of MS2 spectra, and utilizes their experimental MS2 spectra as surrogate spectra to annotate their reaction-paired neighbor metabolites, which subsequently serve as the basis for recursive analysis. Using different LC-MS platforms, data acquisition methods, and biological samples, we showcase the utility and versatility of MetDNA and demonstrate that about 2,000 metabolites can cumulatively be annotated from one experiment. Our results demonstrate that MetDNA substantially expands metabolite annotation, enabling quantitative assessment of metabolic pathways and facilitating integrative multi-omics analysis.
Web Link of MetDNA: http://metdna.zhulab.cn
Publications:
MetDNA: X. Shen, R. Wang, X. Xiong, Y. Yin, Y. Cai, Z. Ma, N. Liu, and Z.-J. Zhu*,Metabolic Reaction Network-based Recursive Metabolite Annotation for Untargeted Metabolomics, Nature Communications, 2019, 10: 1516. Web Link
MetDNA2: Z. Zhou†, M. Luo†, H. Zhang, Y.Yin, Y. Cai, and Z.-J. Zhu* , Metabolite Annotation from Knowns to Unknowns through Knowledge-guided Multi-layer Metabolic Networking, Nature Communications, 2022, 13: 6656. Web Link