
Lipid4DAnalyzer: Development of a combined strategy for accurate lipid structural identification and quantification in ion-mobililty mass spectrometry based untarget lipis
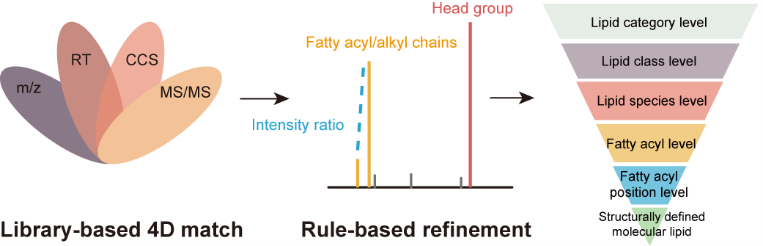
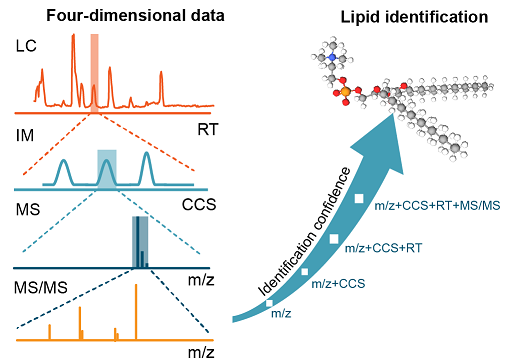
Brief introduction: The lipid identification using the multi-dimensional match (i.e., MS1, retention time, collision cross section, and MS/MS spectra) gives multiple lipid candidates, and often over-reports the structural information. Here, we developed a lipid identification strategy that integrated library-based match and rule-based refinement for accurate lipid structural elucidation in IM-MS based lipidomics. We added the function of rule-based refinement and upgraded LipidIMMS Analyzer to Lipid4DAnalyzer to support more instrument platforms, including both IM-MS (i.e., Agilent 6560 DTIM-MS, Bruker timsTOF Pro, Waters Synapt G2-Si) and non-IM-MS based instruments (i.e., Sciex TripleTOF 6600).
Availability of Lipid4DAnalyzer: http://lipid4danalyzer.zhulab.cn
LipidIMMS Analyzer: Integrating Multi-dimensional Information to Support Lipid Identification in Ion Mobility–Mass Spectrometry based Lipidomics
Brief introduction: Ion mobility - mass spectrometry (IM-MS) has showed great application potential for lipidomics. However, IM-MS based lipidomics is significantly restricted by the available software for lipid structural identification. Here, we developed a software tool, namely, LipidIMMS Analyzer, to support the accurate identification of lipids in IM-MS. For the first time, the software incorporates a large-scale database covering over 260,000 lipids and four-dimensional structural information for each lipid (i.e., m/z, retention time (RT), collision cross-section (CCS) and MS/MS spectra). Therefore, multi-dimensional information can be readily integrated to support lipid identifications, and significantly improve the coverage and confidence of identification. Currently, the software supports different IM-MS instruments and data acquisition approaches.
Availability of LipidIMMS: http://imms.zhulab.cn/LipidIMMS (LipidIMMS has been upgraded to Lipid4DAnalyzer)
Tutorial: Download
FAQs: Download
Demo Data: Download
Publications:
1. X. Chen†, Y. Yin†, M. Luo, Z. Zhou, Y. Cai*, and Z.-J. Zhu* (Corresponding author), Trapped Ion Mobility Spectrometry-Mass Spectrometry Improves the Coverage and Accuracy of Four-dimensional Untargeted Lipidomics, Analytica Chimica Acta, 2022, 1210: 339886.
2. X. Chen†, Y. Yin†, Z. Zhou, T. Li, and Z.-J. Zhu* (corresponding author), Development of A Combined Strategy for Accurate Lipid Structural Identification and Quantification in Ion-Mobility Mass Spectrometry based Untargeted Lipidomics, Analytica Chimica Acta, 2020, 1136, 115-124.
3. X. Chen, Z. Zhou, and Z.-J. Zhu* (Corresponding author), The Use of LipidIMMS Analyzer for Lipid Identification in Ion Mobility-Mass Spectrometry-Based Untargeted Lipidomics, Methods in Molecular Biology, 2020, 2084, 269-282.
4. J. Tu†, Z. Zhou†, T. Li†, and Z.-J. Zhu* (Corresponding Author), The Emerging Role of Ion Mobility-Mass Spectrometry in Lipidomics to Facilitate Lipid Separation and Identification, Trends in Analytical Chemistry, 2019, 116, 332-339.
5. Z. Zhou, X. Shen, X. Chen, J. Tu, X. Xiong, and Z.-J. Zhu* (Corresponding author), LipidIMMS Analyzer: Integrating Multi-dimensional Information to Support Lipid Identification in Ion Mobility–Mass Spectrometry based Lipidomics, Bioinformatics, 2019, 35,698-700.
6. Z. Zhou, J. Tu, X. Xiong, X. Shen, and Z.-J. Zhu* (Corresponding author), LipidCCS: Prediction of Collision Cross-Section Values for Lipids with High Precision to Support Ion Mobility-Mass Spectrometry based Lipidomics, Analytical Chemistry, 2017, 89, 9559–9566.
7. J. Tu, Y. Yin, M. Xu, R. Wang, and Z.-J. Zhu*(Corresponding author), Absolute Quantitative Lipidomics Reveals Lipidome-Wide Alterations in Aging Brain, Metabolomics, 2018,14: 5.