
DecoMetDIA: Deconvolution of Multiplexed MS/MS Spectra for Metabolite Identification in SWATH-MS based Untargeted Metabolomics
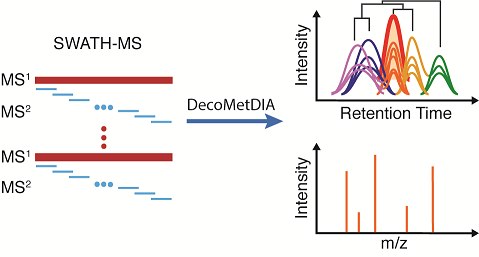
Brief Introduction: SWATH-MS based data independent acquisition mass spectrometry (DIA-MS) technology has been recently developed for untargeted metabolomics due to its capability to acquire all MS2 spectra and high quantitative accuracy. However, software tools for deconvolving multiplexed MS/MS spectra from SWATH-MS with high efficiency and quality are still lacking in untargeted metabolomics. Here, we developed a new software tool, namely, DecoMetDIA, to deconvolve multiplex MS/MS spectra for metabolite identification and support the SWATH based untargeted metabolomics. In DecoMetDIA, it selected multiple model peaks to model the co-eluted and unresolved chromatographic peaks of fragment ions in multiplexed spectra, and decomposed them into a linear combination of the model peaks. DecoMetDIA enabled to reconstruct the MS2 spectra of metabolites from a variety of different biological samples with a high coverage. We also demonstrated that deconvolved MS2 spectra from DecoMetDIA were of high accuracy through the comparison to the experimental MS2 spectra from data dependent acquisition (DDA). Finally, about 90% of deconvolved MS2 spectra in various biological samples were successfully annotated using annotation software tools such as MetDNA and Sirius. This proved that the deconvolved MS2 spectra obtained from DecoMetDIA were accurate and valid for metabolite identification and structural elucidation.
Availability: https://github.com/ZhuMSLab/DecoMetDIA
MetDIA: Targeted Metabolite Extraction of Multiplexed MS/MS Spectra Generated by Data-Independent Acquisition
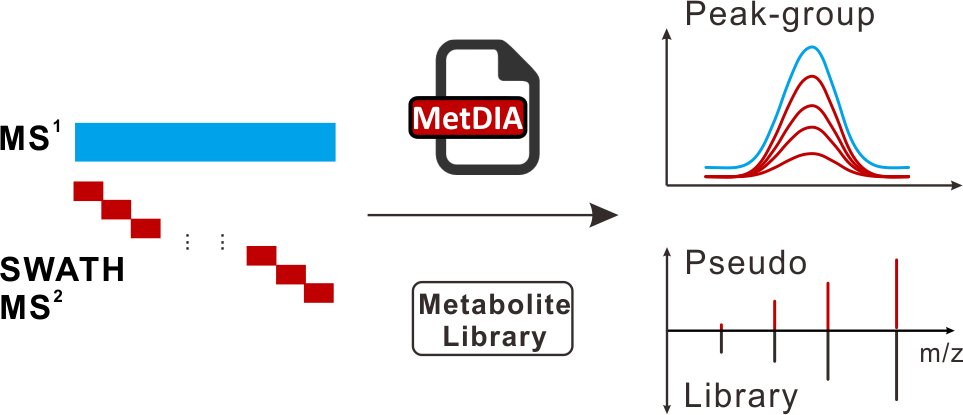
Brief introduction: With recent advances in mass spectrometry, there is an increased interest in data-independent acquisition (DIA) techniques for metabolomics. With DIA technique, all metabolite ions are sequentially selected and isolated using a wide window to generate multiplexed MS/MS spectra. Therefore, DIA strategy enables a continuous and unbiased acquisition of all metabolites and increases the data dimensionality, but presents a challenge to data analysis due to the loss of the direct link between precursor ion and fragment ions. Here, we developed a new metabolite-centric approach, namely, MetDIA, to process DIA data. The comparisons of our MetDIA method with data-dependent acquisition (DDA) method demonstrated that MetDIA could significantly detect more metabolites in biological samples, and is more accurate and sensitive for metabolite identifications. The MetDIA program and the metabolite spectral library is freely available on the Internet.
Download link
MetDIA instruction and demo report
SWATHtoMRM: Development of High-Coverage Targeted Metabolomics Method Using SWATH Technology for Biomarker Discovery
...
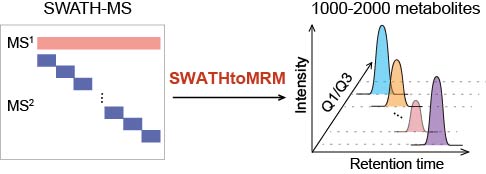
Brief introduction: The complexity of metabolome presents a great analytical challenge for quantitative metabolite profiling, and restricts the application of metabolomics for biomarker discovery. Targeted metabolomics using multiple-reaction monitoring (MRM) technique has excellent capability for quantitative analysis, but is severely restricted by metabolite coverage in one analysis. To address this challenge, we developed a new strategy, namely, SWATHtoMRM, which utilizes the broad coverage of SWATH-MS technology to develop high-coverage targeted metabolomics method. Specifically, SWATH-MS technique was first utilized to untargeted profile the one pooled sample, and to acquire the MS2 spectra for all metabolites. Then, SWATHtoMRM was used to extract large-scale MRM transitions for targeted analysis with a coverage as high as 1000-2000 metabolites in one experiment. The qualification performances of SWATHtoMRM method including good reproducibility, high sensitivity, and broad dynamic range were systematically evaluated and demonstrated. SWATHtoMRM offers a new approach to develop high-throughput targeted metabolomics method and further facilitates discovering potential reliable biomarkers for diseases benefitting from comprehensive metabolites profiling and accurate quantification.
Download link
Binary package: SWATHtoMRM_1.0.0.zip
Source package: SWATHtoMRM_1.0.0.tar.gz
SWATHtoMRM instruction and demo report
Please send email to jiangzhu AT sioc.ac.cn to request a copy of DecoMetDIA/MetDIA/SWATHtoMRM
Publications:
1. Y. Yin†, R. Wang †, Y. Cai, Z. Wang, and Z.-J. Zhu* (Corresponding Author), DecoMetDIA: Deconvolution of Multiplexed MS/MS Spectra for Metabolite Identification in SWATH-MS based Untargeted Metabolomics,Analytical Chemistry, 2019, 91, 11897-11904.
2. H. Li, Y. Cai, Y. Guo, F. Chen, and Z.-J. Zhu*(Corresponding author), MetDIA: Targeted Metabolite Extraction of Multiplexed MS/MS Spectra Generated by Data-Independent Acquisition, Analytical Chemistry, 2016. Web Link
3. H. Zha†, Y. Cai†, Y. Yin†, Z. Wang, K. Li, and Z.-J. Zhu* (Corresponding Author), SWATHtoMRM: Development of High-Coverage Targeted Metabolomics Method Using SWATH Technology for Biomarker Discovery, Analytical Chemistry, 2018, 90, 4062-4070. Web Link (†, Co-first authors)