
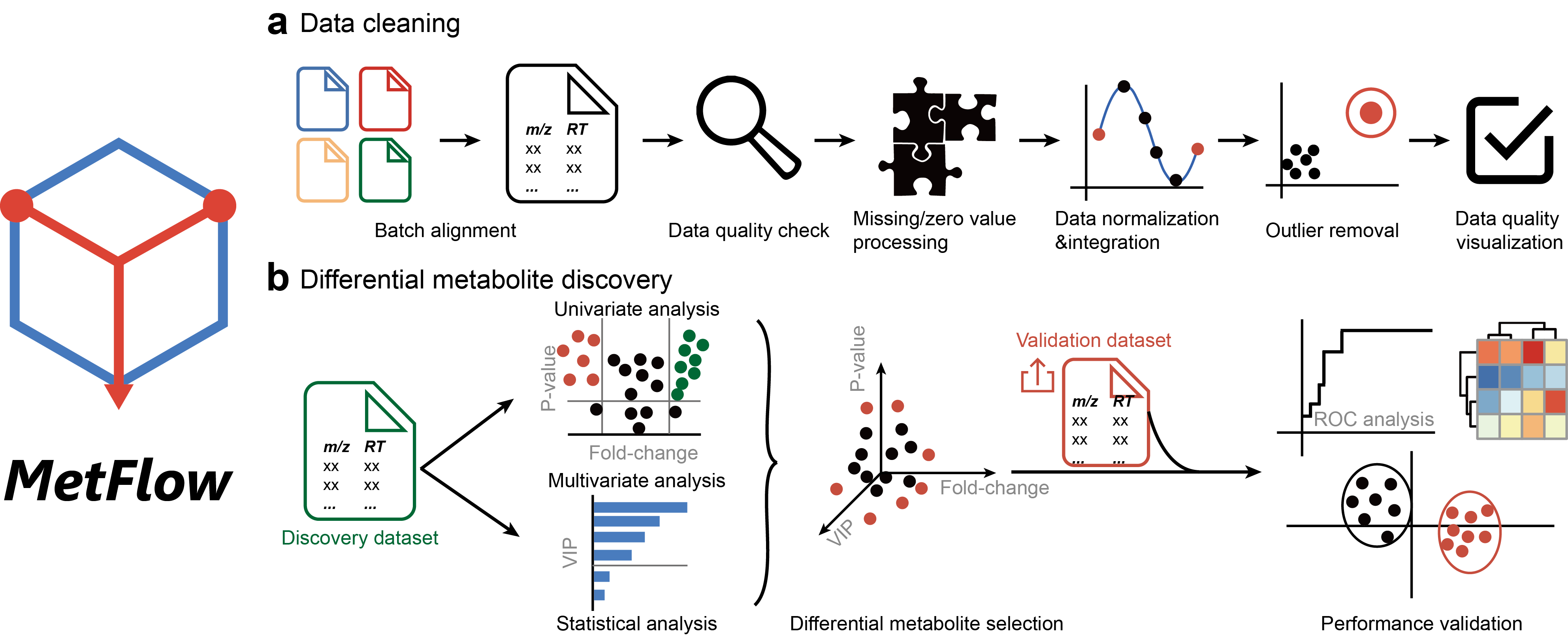
MetFlow: An interactive and integrated workflow for metabolomics data cleaning and differential metabolite discovery
Brief Introduction: Mass spectrometry-based metabolomics aims to profile the metabolic changes in biological systems and identify differential metabolites in relation to physiological phenotypes and aberrant activities. However, many confounding factors during data acquisition complicate metabolomics data, which is characterized by high dimensionality, uncertain degrees of missing and zero values, non-linearity, unwanted variations, and non-normality. Therefore, prior to differential metabolite discovery analysis, various types of data cleaning such as batch alignment, missing value imputation, data normalization and scaling are essentially required for data post-processing. Here, we developed an interactive web server, namely, MetFlow, to provide an integrated and comprehensive workflow for metabolomics data cleaning and differential metabolite discovery.
Availability of MetFlow: http://metflow.zhulab.cn
MetNormalizer: Normalization and Integration of Large-Scale Metabolomics Data Using Support Vector Regression
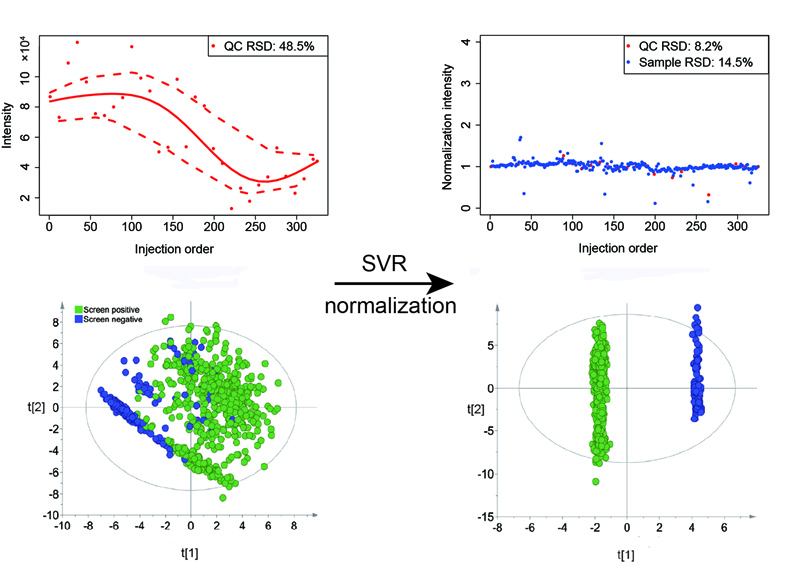
Brief Introduction: Untargeted metabolomics studies for biomarker discovery often have hundreds to thousands of human samples. Data acquisition of large-scale samples has to be divided into several batches and may span from months to as long as several years. The signal intensity drift of metabolites during data acquisition (intra- and inter-batch) is unavoidable and is a major confounding factor for large-scale metabolomics studies. Therefore, the combination of multiple batches in large-scale metabolomics studies requires proper data normalization to reduce unwanted variations prior to statistical analyses. Here, we developed a machine learning algorithm-based method, support vector regression (SVR), for large-scale metabolomics data normalization and integration. The unwanted intra- and inter-batch variations can be effectively removed after SVR normalization. We demonstrated that the portion of metabolic peaks with RSDs less than 30% increased to more than 90% of the total peaks after SVR normalization, which is much better than other common normalization methods. The reduction of unwanted analytical variations helps improve the performance of multivariate statistical analyses, both unsupervised and supervised, in terms of classification and prediction accuracy so that subtle metabolic changes in epidemiological studies can be detected. An R package named MetNormalizer was developed and provided for data processing using SVR normalization.
Download link:
Update on April, 2016
This work has been selected as ORAL presentation on 64thASMS conference on Mass Spectrometry (June 4-9, 2016 at San Antonio, TX, United States).
Update on August, 2016 by X. Shen
The newest version of MetNormalizer can be installed using github (http://github.com/jaspershen/MetNormalizer).
(1) install.packages("devtools")
(2) library(devtools)
(3) install_github("jaspershen/MetNormalizer")
(4) library(MetNormalizer)
Publications:
1. X. Shen, and Z.-J. Zhu* (Corresponding author), MetFlow: An Interactive and Integrated Workflow for Metabolomics Data Cleaning and Differential Metabolite Discovery, Bioinformatics, 2019, 35, 2870-2872. Web Link Web Server Link
2. X. Shen, X. Gong, Y. Cai, Y. Guo, J. Tu, H. Li, T. Zhang, J. Wang, F. Xue, and Z.-J. Zhu* (corresponding author), Normalization and Integration of Large-Scale Metabolomics Data Using Support Vector Regression, Metabolomics, 2016,12: 89 Web Link